Archives
- 2026-02
- 2026-01
- 2025-12
- 2025-11
- 2025-10
- 2025-09
- 2025-03
- 2025-02
- 2025-01
- 2024-12
- 2024-11
- 2024-10
- 2024-09
- 2024-08
- 2024-07
- 2024-06
- 2024-05
- 2024-04
- 2024-03
- 2024-02
- 2024-01
- 2023-12
- 2023-11
- 2023-10
- 2023-09
- 2023-08
- 2023-07
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
- 2018-11
- 2018-10
- 2018-07
-
CCT251545 receptor br Materials and methods br Results
2019-12-17
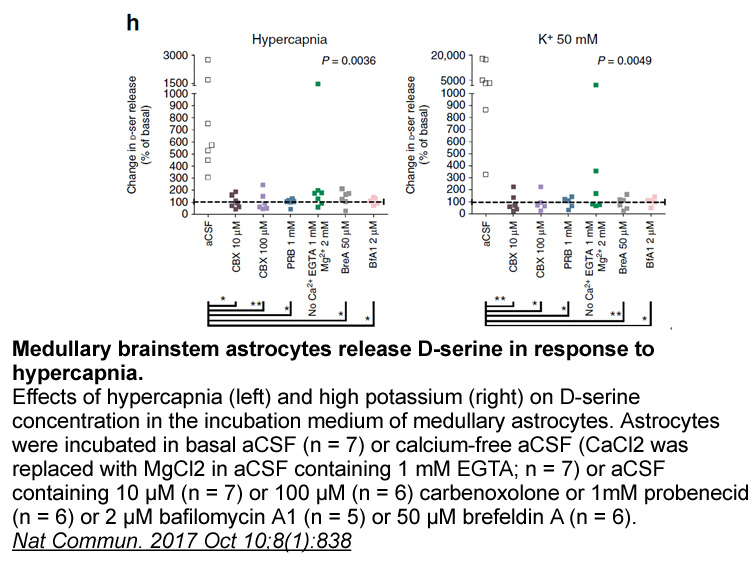
Materials and methods Results Discussion In the past decade, GWAS have attempted to identify genetic variants that confer risk for many human diseases, whose inherited components remain unexplained (Manolio et al., 2009). In a few cases risk variants identified by GWAS have paved the way fo
-
In this paper we report the synthesis and microbiological ev
2019-12-17
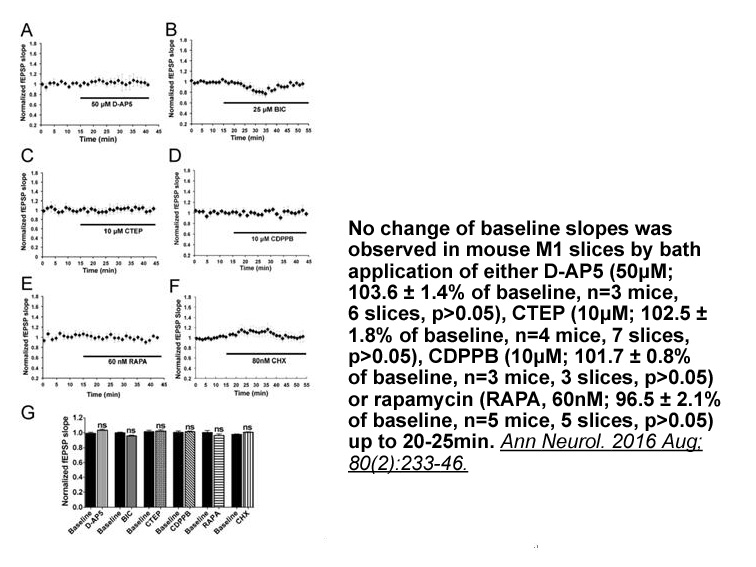
In this paper, we report the synthesis and microbiological evaluation of a series of novel chromogenic sugar-based enzyme substrates based upon catechol, 2,3-dihydroxynaphthalene and 6,7-dibromo-2,3-dihydroxynaphthalene 9 cores (Fig. 1).14, 15 2,3-Dihydroxynaphthalene is inexpensive and available in
-
br Acknowledgments This work was supported by R gion Auvergn
2019-12-17
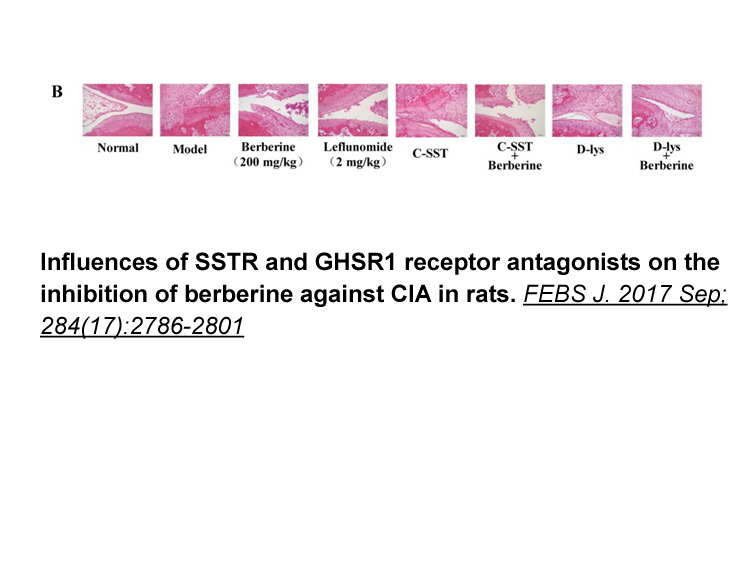
Acknowledgments This work was supported by Région Auvergne “Nouveau chercheur”. This study has been performed with the assistance of Christelle Damon-Soubeyrand for histology technical assistance using “Anipath” Platform (GReD) and Sandrine Plantade, Keredine Ouchen and Philippe Mazuel for animal
-
br Materials and Methods br Acknowledgements Authors would l
2019-12-17
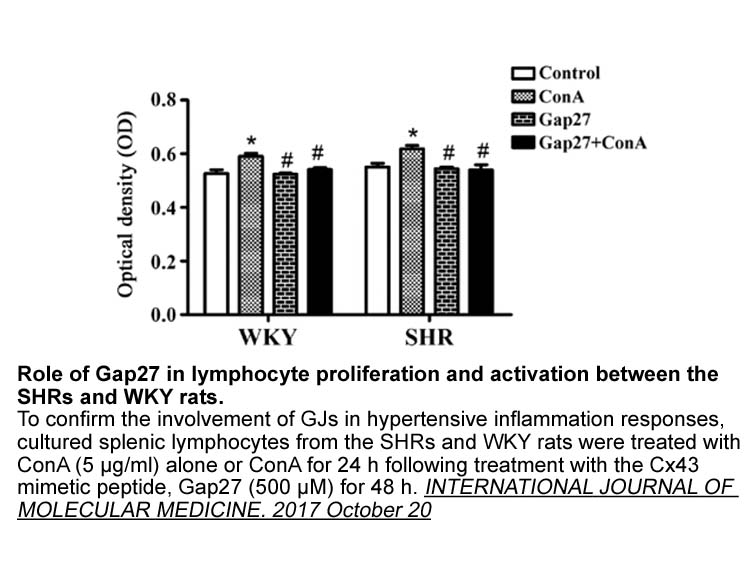
Materials and Methods Acknowledgements Authors would like to thank Mr. Pritam Naskar and Mr. Dibya Mukherjee for their help. Authors also acknowledge the help of Mr. Barun Mahata and Dr. Kaushik Biswas, Division of Molecular Medicine, Bose Institute, for human cDNA samples. P.A.B. and A.B.D. w
-
Plants Cp are widely distributed in
2019-12-17
Plants Cp are widely distributed in the plant kingdom and are believed to act as virulence/defense factors for both hosts and pathogens. Cp are also found in plants, animals and bacteria and are known to be virulence factors involved in bacterial pathogenicity [29]. Further, Cp are involved in pepti
-
LY2874455 Instead of introducing extra hardware or hardware
2019-12-17
Instead of introducing extra hardware or hardware modification, software-based CFC techniques just insert extra checking instructions into the source code of the target program at compile time so as to have the target program do checking jobs itself. Preeminent software-based CFC techniques proposed
-
In addition to PGC transcriptional co repressors
2019-12-17
In addition to PGC1α, transcriptional co-repressors such as NCOR and RIP140 participate in oxidative muscle remodeling induced by exercise, whereby reductions in their expression and the resulting de-repression of downstream TFs activates oxidative gene expression (Seth et al., 2007, Yamamoto et al.
-
br Materials and methods br Results br Discussion
2019-12-17

Materials and methods Results Discussion Our previous study showed that ERRγ negatively regulates osteoblast differentiation via inhibiting Runx2 transactivity (Jeong et al., 2009). In the present study, we found that ERRγ also induced miR-433 in osteogenic mesenchymal cell lineage C3H10T1/
-
br Introduction Epidermal growth factor
2019-12-17
Introduction Epidermal growth factor receptor (EGFR)-activating-mutant non-small cell lung cancer (NSCLC) often initially responds well to EGFR tyrosine kinase inhibitors (TKIs) (Haber et al., 2011); however, the disease almost always recurs about 10–13 months of therapy. Analysis of clinical spe
-
ap1 br Results br Discussion Our UbV
2019-12-17
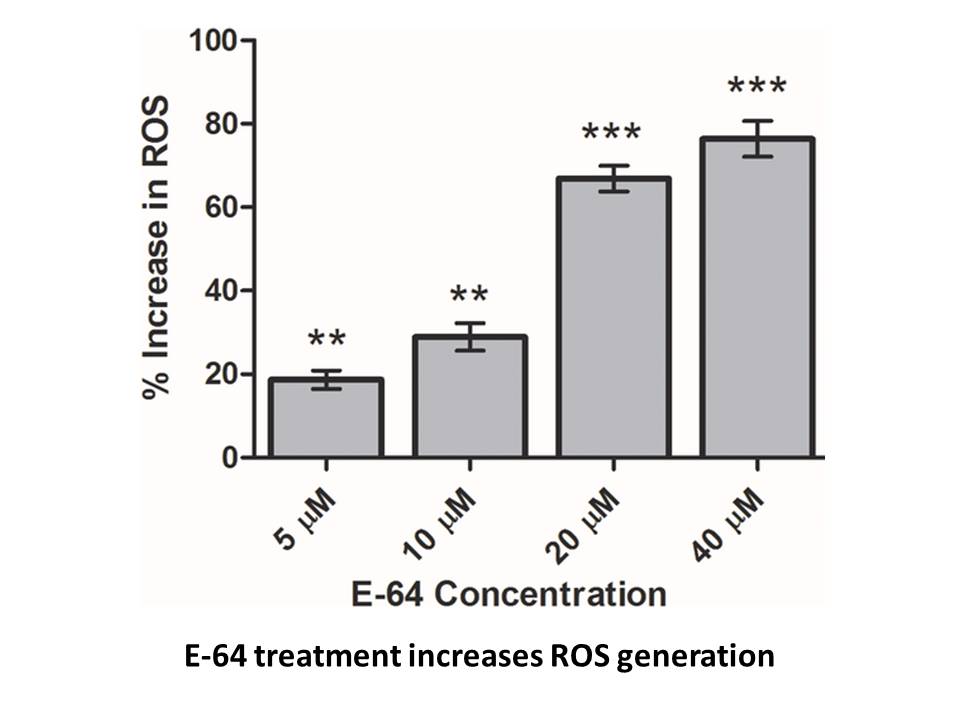
Results Discussion Our UbV library was originally designed to develop inhibitors of deubiquitinases (Ernst et al., 2013). Recently, we showed that UbVs could exhibit multiple binding modes and mechanisms to modulate HECT E3 activity (Zhang et al., 2016)—one set occupied the HECT domain E2-bind
-
auz br Materials and methods br Results br Discussion COX me
2019-12-16
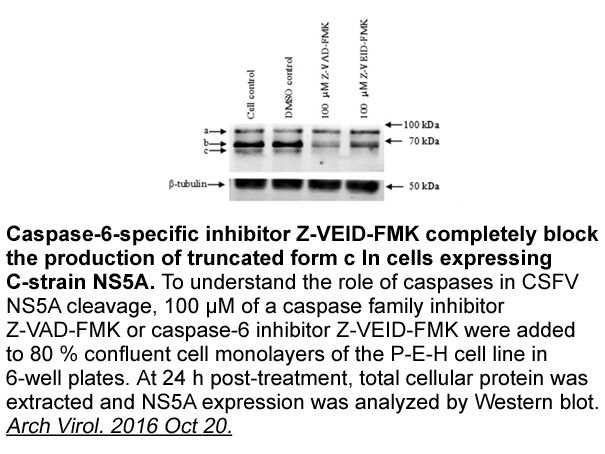
Materials and methods Results Discussion COX-2-mediated production of PGE2 is involved in cell growth and metastasis of many cancers. Previous studies indicated that COX-2 was overexpressed in many cancer tissues and that PGE2 increased cancer cell growth, a process that could be suppressed
-
br Disclosures br Acknowledgements This
2019-12-16
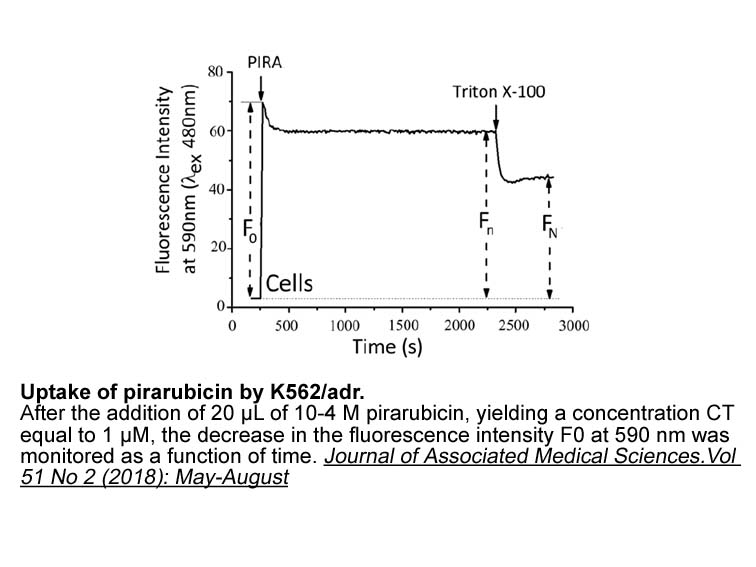
Disclosures Acknowledgements This work was supported by the Natural Science Foundation of China (No. 81560587); the Fok Ying-Tong Education Foundation of China (No. 151107), the basic ability promotion project for young and middle-aged teachers in Universities in Guangxi (No. 2018KY1146) and t
-
In contrast to SQLE HMGCR could be efficiently degraded in
2019-12-16

In contrast to SQLE, HMGCR could be efficiently degraded in Imipramine hydrochloride lacking UBE2J2. However, this was not the case in cells devoid of UBE2G2, as these cells were unable to support 25-hydroxycholesterol (25-HC)-stimulated degradation of HMGCR (Fig. 1C). This finding is consistent wi
-
br Experimental Procedures br Author Contributions M B
2019-12-16
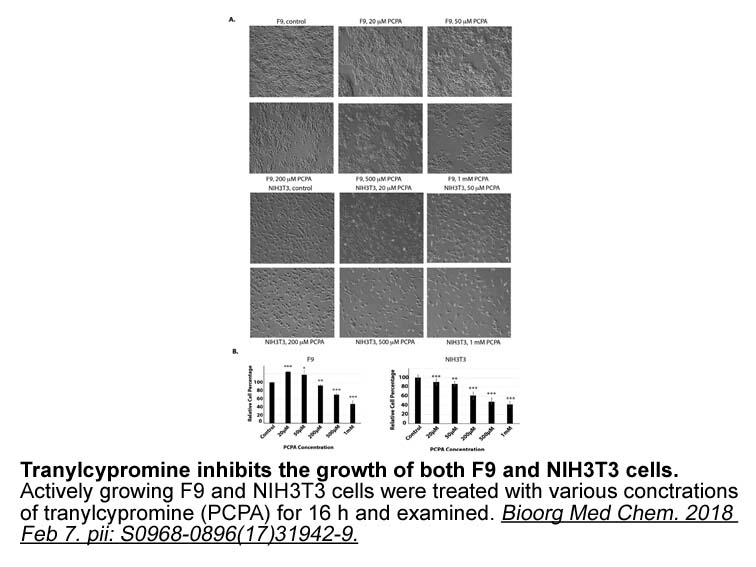
Experimental Procedures Author Contributions M.B., with input and help from T.T.H., conceived the project, designed and performed the experiments, analyzed the data, and wrote the manuscript. B.B.B. and F.D.M. synthesized the wild-type and (nonhydrolyzable, NH) Ub tetramers and contributed Fig
-
Mitochondrial DNA Isolation Kit sale In a recent study Tomki
2019-12-16
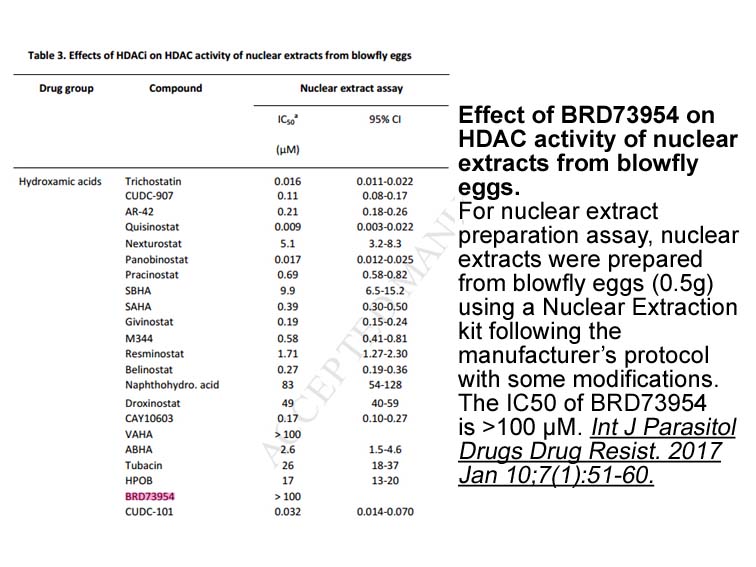
In a recent study, Tomkins et al. [7] analyzed the protein-protein interaction network of ROCO proteins. Based on a database analysis, and only taking into account interactions reported in at least two peer-reviewed papers and/or confirmed by two different methods, 113 interactors were revealed for
16668 records 957/1112 page Previous Next First page 上5页 956957958959960 下5页 Last page